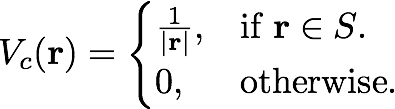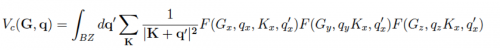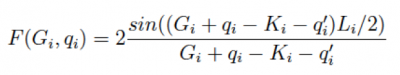How to treat low dimensional systems
In this tutorial you will learn for a low-dimensional (2D) material how to:
- Avoid numerical divergence using the Random Integration Method (RIM)
- Generate a truncated coulomb potential with a box-like cutoff to eliminate the image-image interactions
- Use the truncated coulomb potential in the GW calculation
- Use the truncated coulomb potential in the BSE calculation
- Analyze the difference with corresponding calculations without the use of a truncated potetnial
Prerequisites
- Complete the Generating the Yambo databases tutorial
SAVEfolder for 2D hBN.yamboexecutableyppexecutable- Run Initialization
Avoid numerical divergence using the Random Integration Method
In a low-dimensional material, like 2D-hBN, less than 3D k-grids are generally used i.e. NxNx1 This can create a numerical instability of the q-space integration due to the q=0 part of the Coulomb Potential in all the main equations (see i.e. the exchange self-energy equation) if a dense sampling is used.
To eliminate this problem YAMBO uses the so-called Random Integration Method which means to use a Monte Carlo integration with random Q-points whose number RandQpts is given in input.
Create the input to generate the ndb.RIM database
$ yambo -F yambo_RIM.in -r
RandQpts= 1000000 # [RIM] Number of random q-points in the BZ RandGvec= 1 RL # [RIM] Coulomb interaction RS components
Close input and Run yambo
$ yambo -F yambo_RIM.in -J 2D
At the end in the 2D directory you will have a new database
ndb.RIM
The presence of this numerical problems becomes evident using denser k-grid with respect to that used in this Tutorial (6x6x1).
Generating new SAVE for 15x15x1 and 30x030x1 k-grids and performing HF calculations you will see the problem!
So home message : use always the RIM in MB simulations of low-dimensional materials.
Here we report the HF gap calculated with the 3 k-grids without (noRIM) and with Random Inetgration Method (RIM)
noRIM RIM 6x6x1 15x15x1 30x30x1
Generate a truncated coulomb potential/ndb.cutoff database (yambo -r)
To simulate an isolated nano-material a convergence with cell vacuum size is in principle required, like in the DFT runs. The use of a truncated Coulomb potential allows to achieve faster convergence eliminating the interaction between the repeated images along the non-periodic direction (see i.e. D. Varsano et al Phys. Rev. B and .. ) In this tutorial we learn how to generate a box-like cutoff for a 2D system with the non-periodic direction along z.
In YAMBO you can use :
spherical cutoff (for 0D systems) cylindrical cutoff (for 1D systems) box-like cutoff (for 0D, 1D and 2D systems)
The Coulomb potential with a box-like cutoff is defined as
Then the FT component is
where
For a 2D-system with non period direction along z-axis we have
Important remarks:
- the Random Integration Method (RIM) is required to perform the Q-space integration
- for sufficiently large supercells a choose L_i slightly smaller than the cell size in the i-direction ensures to avoid interaction between replicas
Creation of the input file:
$ yambo -F yambo_cut2D.in -r
Open the input file yambo_cut2D.in
Change the variables inside as:
RandQpts= 1000000 # [RIM] Number of random q-points in the BZ RandGvec= 100 RL # [RIM] Coulomb interaction RS components
CUTGeo= "box z" # [CUT] Coulomb Cutoff geometry: box/cylinder/sphere X/Y/Z/XY.. % CUTBox 0.00 | 0.00 | 32.0 | # [CUT] [au] Box sides
Close the input file
Run yambo:
$ yambo -F yambo_cut2D.in -J 2D
in the directory 2D you will find the two new databases
ndb.RIM ndb.cutoff